Obtain Molecule xyz Files from Crystal¶
The information about the individual molecules obtained from the ACSD run command are contained in the crystal xyz file. However, it is sometimes convenient to work with or view the molecules from the crystal individually as individual xyz files.
We will use the SUMELF get_molecules module to obtain the xyz files of the individual molecules of a crystal. This modules will look through the crystal_database folder containing all the xyz files that you obtained from the CCDC using the ACSD program, and convert them into xyz files.
To run this module, run the following in a terminal:
# First, change directory into the path containing the "crystal_database" folder
cd path_to_directory_containing_the_crystal_database_folder
# Second, run the "SUMELF get_molecules" module
SUMELF get_molecules
This will import all the crystal xyz files from the crystal_database folder, extract the individual molecules from the crystals, and save the xyz files of the molecules into a folder called crystal_database_molecules.
You can view the molecules on your favourite xyz viewer. Mine is ase gui. Click here to learn more about how to use ase gui.
Other options¶
There are several other options that you can give to the SUMELF get_molecules command:
--crystal_database_foldername: This is the folder containing the crystal structures you want to separate into individual molecules. The default for this iscrystal_database.--output_molecules_crystal_database_foldername: This is the folder that thexyzfiles of molecules will be written to. The default for this iscrystal_database_molecules.
An example of using these inputs are given below:
SUMELF get_molecules --crystal_database_foldername=example_name_for_crystal_database --output_molecules_crystal_database_foldername=example_name_for_crystal_database_molecules
Example¶
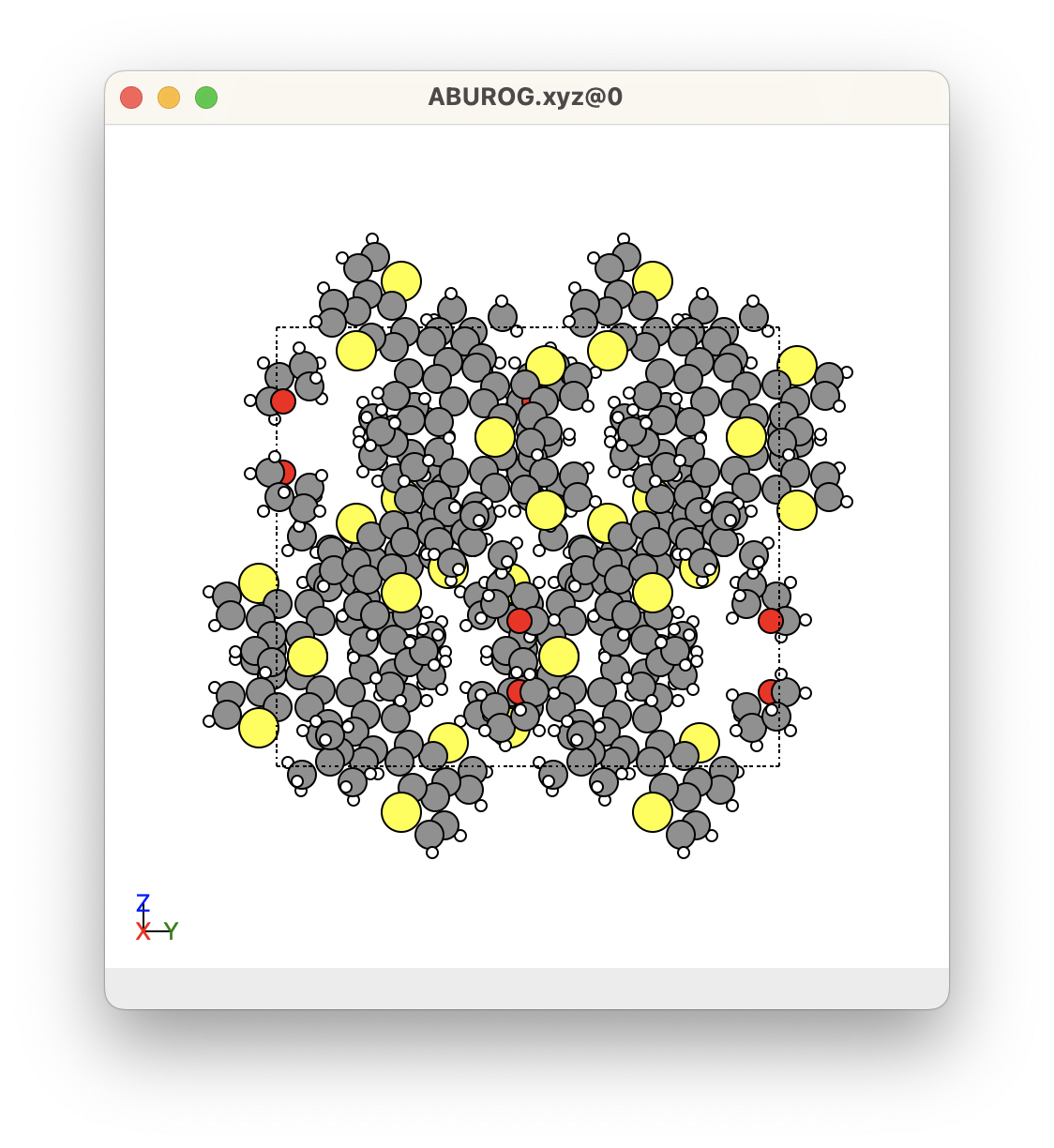
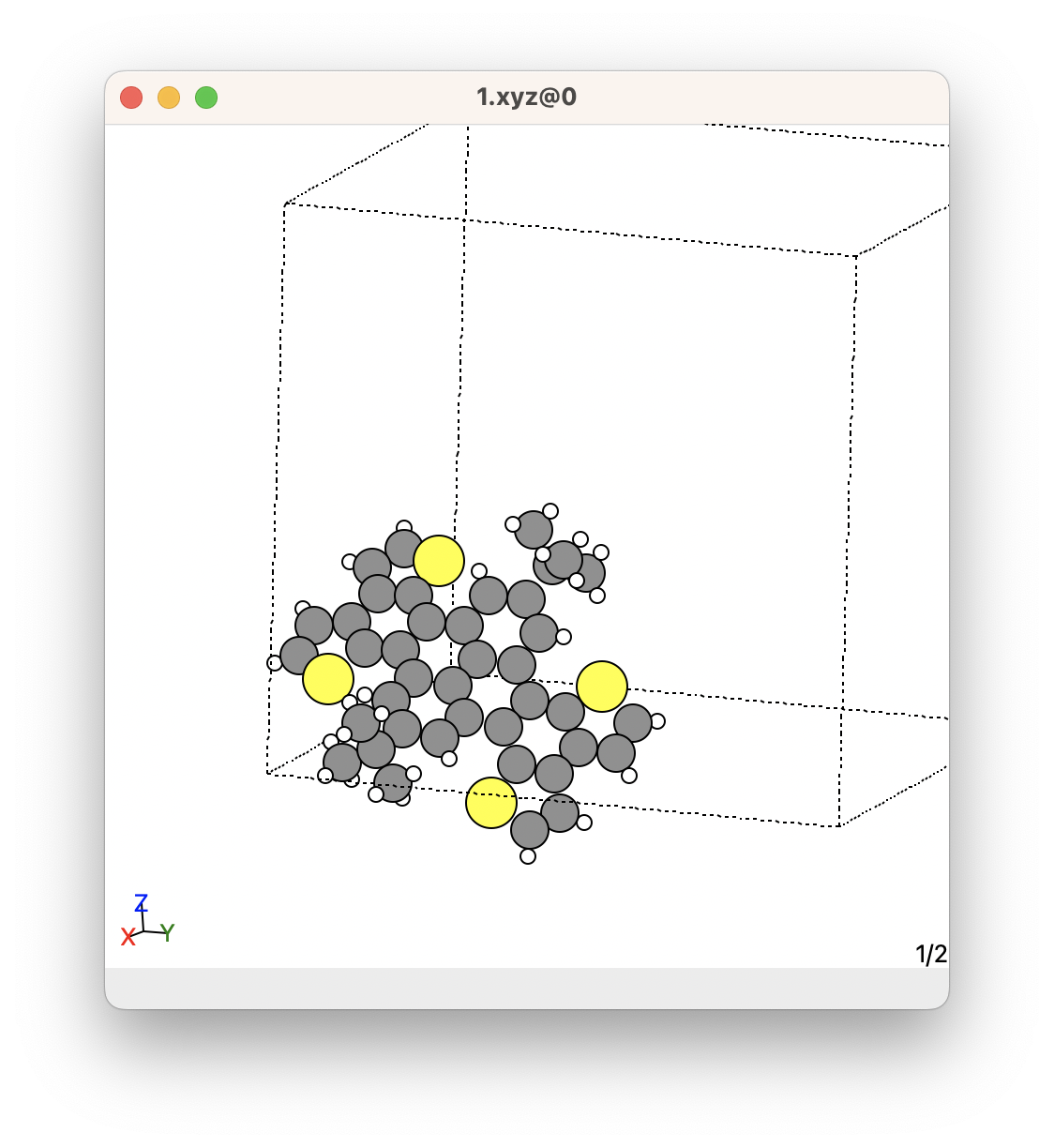
Below shows an example of the xyz files of the molecules from a crystal. In this crystal, we can see that there are many molecules in the unit cell. However, this crystal has a C 2/c space group, so it contains a lot of symmetrically equivalent molecules. There are only two molecules that are unique. These were obtained using the SUMELF get_molecules shown below