Before you Begin - Preliminary Check¶
Before running the RSGC program, it is a good idea to check the crystals you want to remove aliphatic sidechain from are not broken in some way.
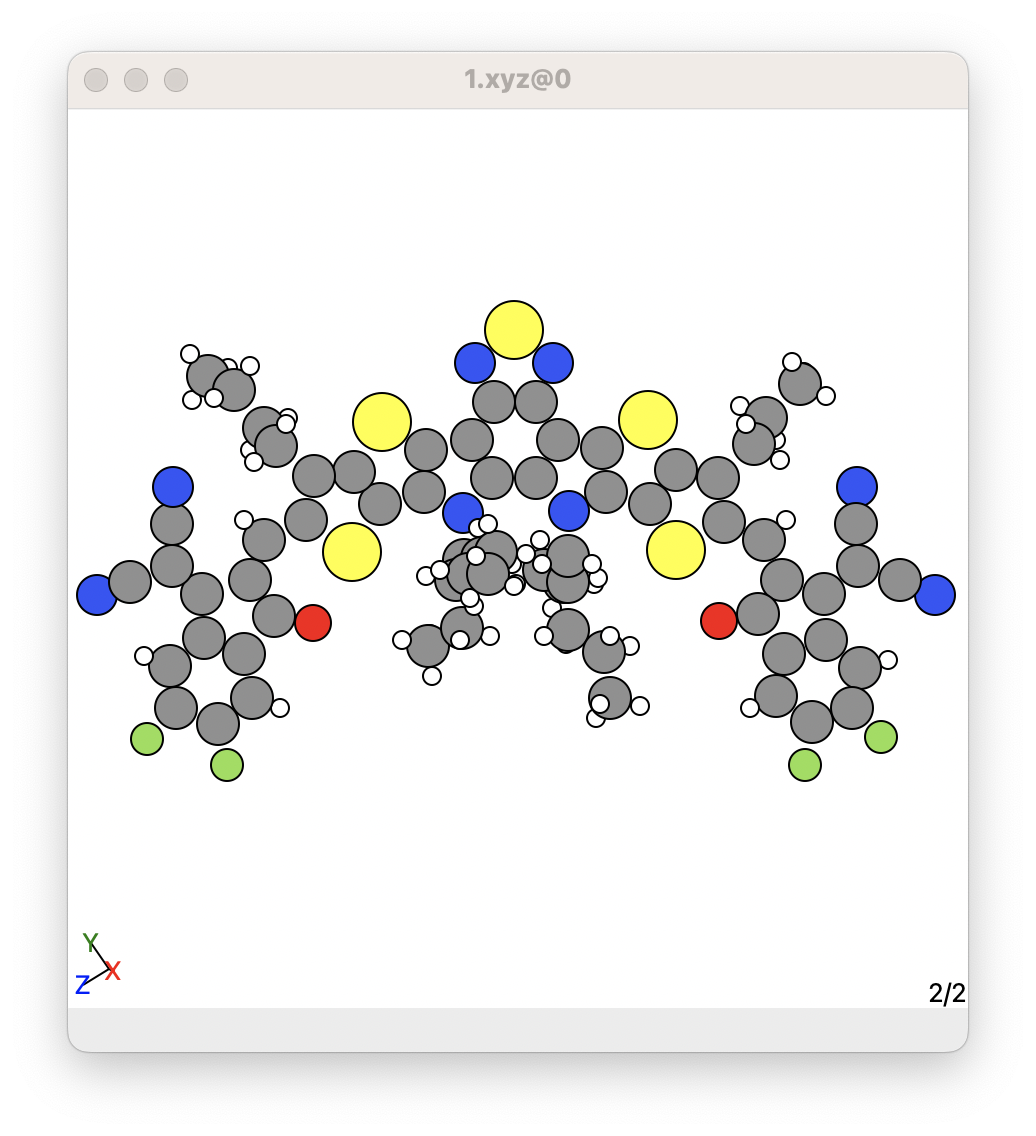
For example, it is common for some crystal structures to have missing hydrogens. An example is shown below for a molecule in the MUPMOC crystal:
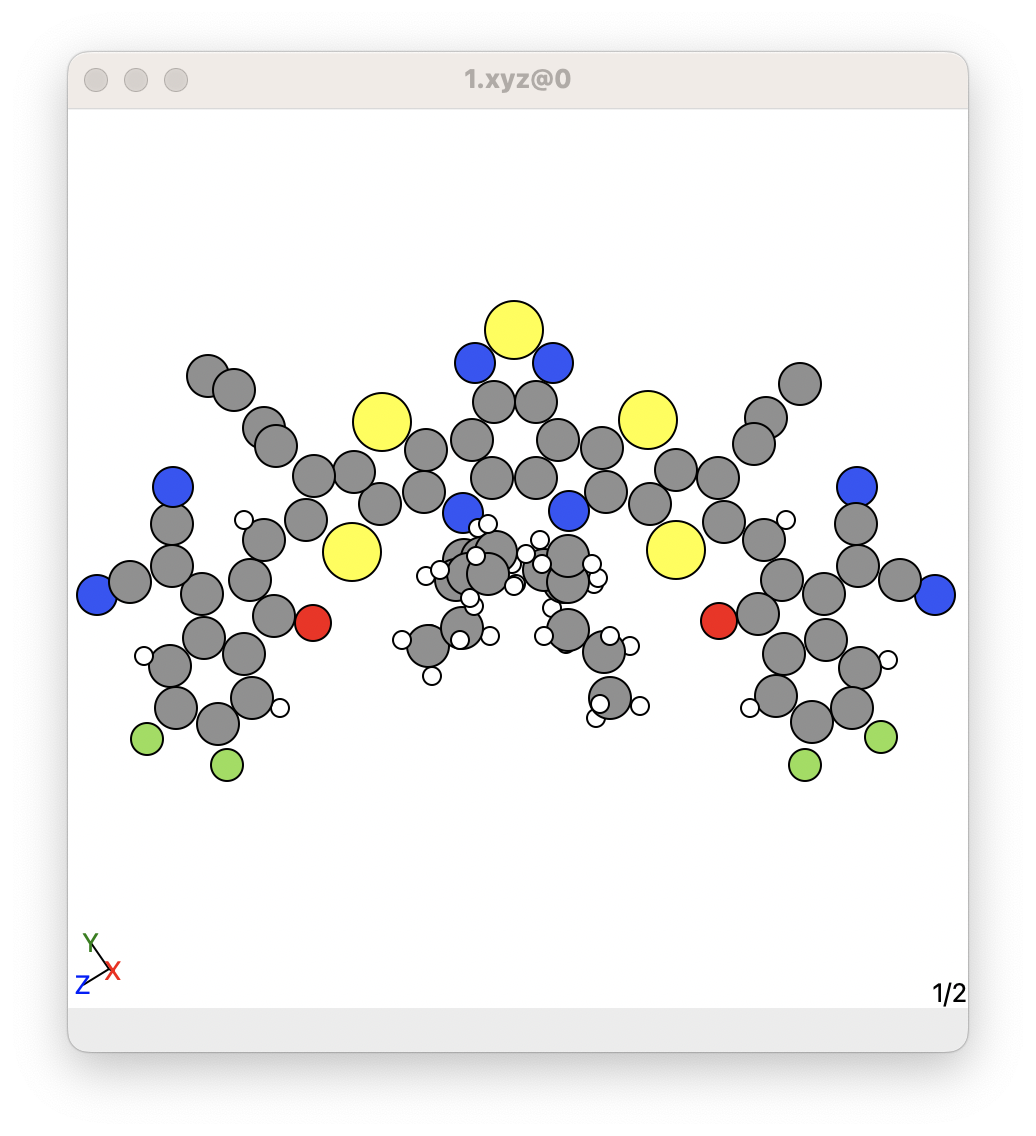
Here, this molecule contains several aliphatic sidechains that are just made up of sp3 carbons and hydrogen atoms. However, some of the hydrogens are missing in these aliphatic sidechains.
It is important not to assume these are fully saturated alipatic sidechain, because maybe there is infact a double bonded or triple bonded carbon sidechain and it is important to included it in your calculations.
- For example, maybe there is a double bond that is important to include in a light absorbing system because it affects the conjugated system, and therefore they affect what the energy levels are of the important molecular orbital energies.
- We could look for the signs that describe a double or triple bond, such as bond lengths and bond angles, however my experience is that in crystal structures sometimes unexpected bond lengths and angles can be found in saturated and unsaturated sidechains
For this reason, the RSGC program does not assume any sidechains with missing hydrogens are saturated.
The best way to fix or repair any molecules for any reason, including missing hydrogens, is to use the ReCrystals program. See the ReCrystals program website for more information. After using this method, you can add hydrogens to these crystal structures, as shown below: